
The plan to release the LGE-LVP dataset
The LGE-LVP dataset is currently under peer review as part of its companion publication. Subject to successful acceptance and final compliance checks, the full dataset is projected to be publicly released by November 2025.
Prompt coronary reperfusion is essential in STEMI to limit infarct size (quantified by percentage of infarcted myocardium) and improve prognosis. However, microvascular obstruction (MVO) persists in 50% of cases despite successful PCI, leading to poor long-term outcomes. Late gadolinium enhancement cardiac MRI (LGE-CMR) serves as the gold standard for MVO detection, enabling precise quantification that strongly predicts adverse outcomes including LV remodeling, heart failure, and mortality. Comprehensive segmentation of LV structures (cavity, myocardium) and pathological features (MVO, infarct) from LGE-CMR provides critical prognostic information, forming the foundation for risk-adapted therapeutic strategies.
Our Contributions
To establish a standardized assessment baseline for MVO and infarct in STEMI patients and promote clinical translation of AI-assisted prognostic models, we release a benchmark dataset segment LV and its pathologies in LGE-CMR images. The details of the dataset are given as follows.
You can download the agreement from here. Please make sure that a permanent/long-term responsible person (e.g., professor) fills in the agreement with a handwriting signature. After filling it, please send the electrical version to our Email: fengchaolu at cse.neu.edu.cn (Subject: LGE-LVP-Agreement). Please send it through an academic or institute email-addresses such as xxx at xxx.edu.xx. Requests from free email addresses (outlook, gmail, qq etc) will be kindly refused. After confirming your information, we will send the download link to you via Email. You need to follow the agreement. Usually we will reply in a week. But sometimes the mail does not arrive and display successfully for some unknown reason. If this happened, please change the content or title and try sending again.
The characteristics of the data,images and patients.
Data Characteristics
Imaging Modality: Cardiac MRI (CMR)
Scanner: 3.0T Philips Ingenia System
Scan Period: Dec 2018 - Sep 2023
Scan Location: Ren Ji Hospital, Shanghai
Contrast Protocol:
Late Gadolinium Enhancement (LGE) imaging
Contrast agent: 0.15 mmol/kg Gd-DTPA (Magnevist, Bayer)
Acquired 10 minutes post-injection
Sequence: 2D Phase-Sensitive Inversion Recovery (PSIR)
Key Parameters:
TR/TE: 6.1/3 ms | Flip angle: 25°
FOV: 300 × 300 mm² | Slice thickness: 10 mm
Resolution: 0.89 × 0.89 mm² (reconstructed)
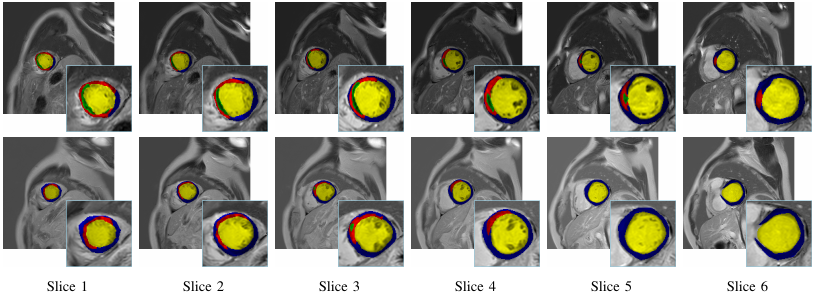
Annotation:Expert-labeled MVO (Microvascular Obstruction), MI(myocardial infarction) myocardium and cavity regions.
Patient Characteristics
Cohort Size: 140 STEMI (ST-segment elevation myocardial infarction) patients
Clinical Context:
All patients underwent emergency PCI (percutaneous coronary intervention)
CMR scans performed ~7 days post-PCI
Inclusion Criteria:
Diagnosed STEMI with PCI history
Complete LGE imaging with identifiable MVO
Ethics: Compliant with the Declaration of Helsinki
The ground truth annotations were generated through a rigorous multi-observer protocol to ensure reliability:
Initial Delineation: Two board-certified radiologists (with 10 years of experience in CMR) independently delineated the regions of interest using MaZda (version 4.6, Institute of Electronics, Technical University of Łódź, Poland; http://www.eletel.p.lodz.pl/mazda/), a validated software tool for medical image analysis.
Consensus Building:After initial annotation, the two observers jointly reviewed all cases to resolve discrepancies through discussion, aiming for a unified annotation set.
Arbitration for Disagreements: In cases where consensus could not be reached (e.g., due to ambiguous anatomical boundaries or imaging artifacts), a senior radiologist with over 20 years of domain expertise was consulted as an arbiter to adjudicate and finalize the GT.
A set of example CMR slices and corresponding annotations is listed as follows.
Researchers can validate their results against our benchmark through the following streamlined process:
Standardized Evaluation Pipeline:We provide an evaluate.py script with pre-implemented metrics (e.g., Dice score, Hausdorff distance).
Usage Example:
bash
python evaluate.py \
--image_dir /path/to/test_images \
--label_dir /path/to/ground_truth \
--pred_dir /path/to/model_predictions
Custom Path Configuration:
Users must manually specify these directory paths:
image_dir: Preprocessed test images (in our released format)
label_dir: Corresponding ground truth annotations
pred_dir: Model prediction files (should match image filename conventions)
Validation Best Practices:
For fair comparison:
Use our official test set split (specified in dataset.json)
Maintain original image resolution (0.89×0.89mm²) and orientation
Advanced Validation Options:
The script supports:
bash
--metrics (-m) # Select specific metrics
--output_csv # Export quantitative results
--visualize # Generate overlay comparisons
More informations are referred in our paper!
Codes can be downloaded in Our Github!

Background
The increased pericardial adipose tissue (PEAT) is associated with a series of cardiovascular diseases (CVDs) and metabolic syndromes. Therefore, particular emphasis should be placed on PEAT quantification and its role in CVD development and progression. In addition, given the more promising clinical value of quantifying PEAT in cardiovascular magnetic resonance (CMR) images than computed tomography (CT) images in CVD assessment, an accurate and time-saving PEAT measurement based on CMR is indispensable.
Motivation
However, almost all existing studies are based on computed tomography (CT) diagnostic modality. Although there is a study segmenting EAT in CMR images in the standard four-chamber(4CH) view, this dataset is unavailable and cardiac short axis (SA) MR images are more prevalent in CVD diagnosis. Segmentation of PEAT in cardiac SA MR images still an pending problem due to the following challenges, 1) the absence of public CMR datasets, 2) diversity in shape, size and location of PEAT, 3) severe class imbalance , and 4) heavily overlapping in intensity.
Our Contributions
To address the above challenges, we release a benchmark dataset and propose a deep learning model to segment PEAT in cardiac SA MR images. The main contributions of our paper are given as follows.
A cardiac SA MR image dataset is released with PEAT being labelled by a radiologist and checked by another and a multiple-stage deep learning model is proposed to segment PEAT in the dataset.
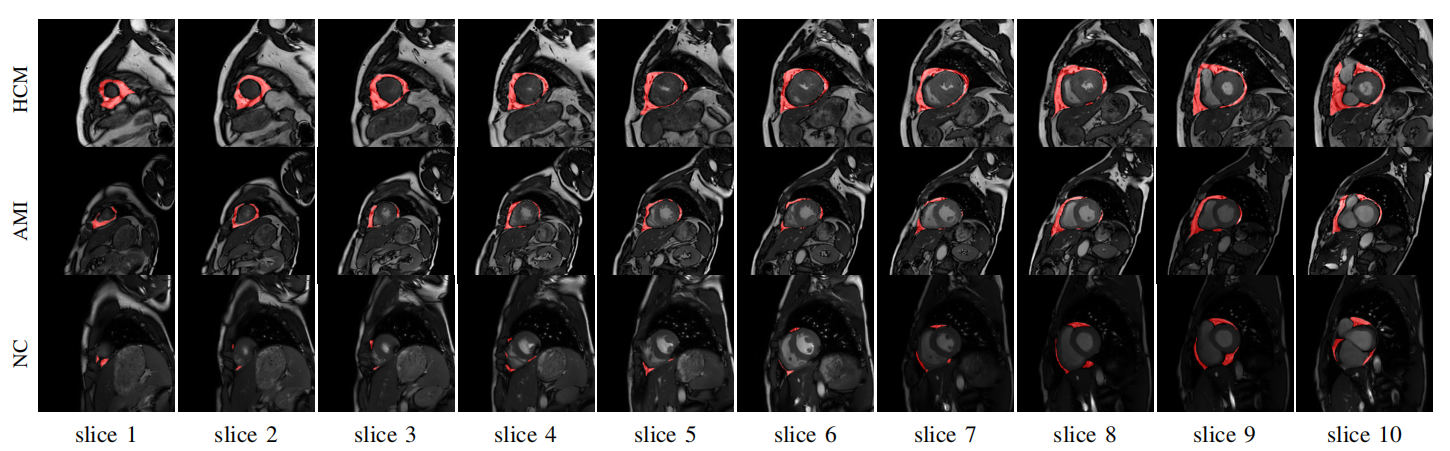
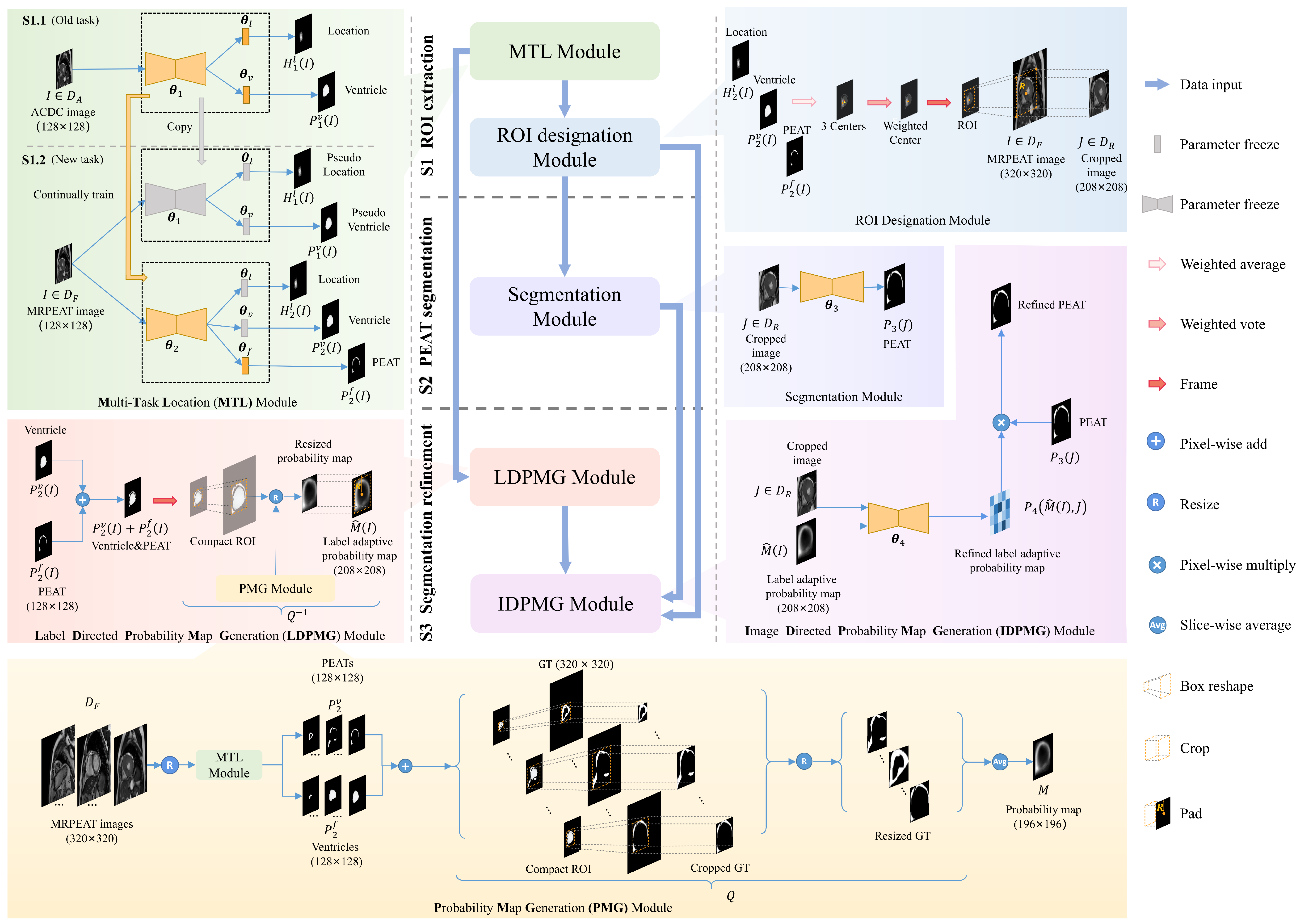
Ventricle segmentation and location are adopted as two additional tasks to assist the model in locating PEAT by ensuring surroundings meet with clinical anatomy.
Continual learning is used to transfer knowledge from existing datasets with cardiac ventricle labels to release additional requirements on labelling our dataset.
A prior guidance strategy is proposed to refine PEAT segmentation results by retrieving knowledge from ground truth labels and image contents.
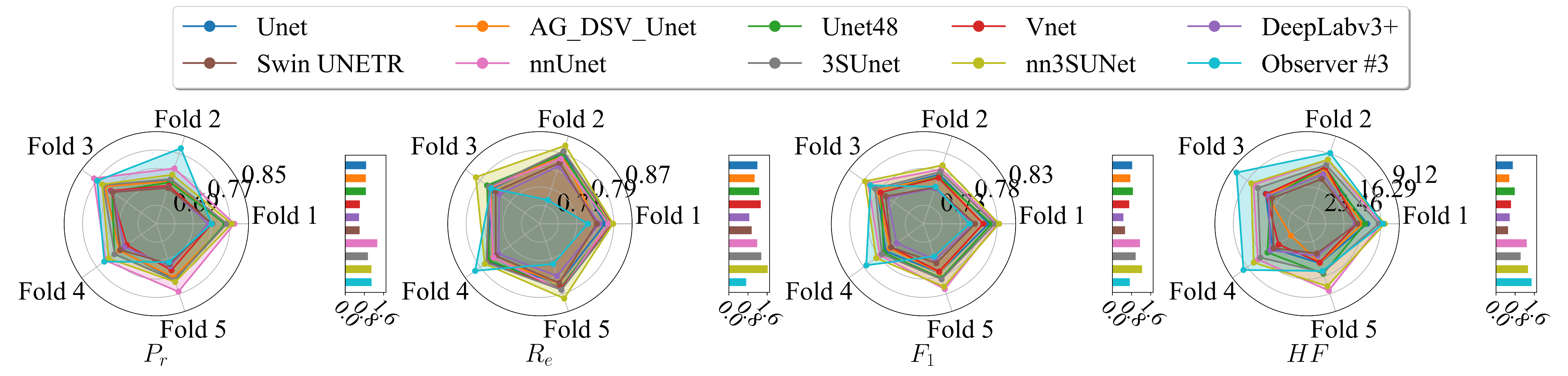
The segmentation model is qualitatively and quantitatively compared with state-of-the-art models on the dataset we released.
More informations are referred in our paper!
Now!
Dataset can be downloaded in Our
!
Codes can be downloaded in Our Github!

Background
Person re-identification aims to retrieve a given person (query) from a large number of candidate images (gallery). Existing deep learning-based methods usually train the model on a fixed scenario (domain). During inference, features of the query and gallery are extracted by the trained model. The similarity between query and gallery features is then measured by Euclidean or cosine distance to match the query from the gallery.
Motivation
However, as the application scenario changes, which is actually the common case, the models will not perform well if it is deployed directly on these changed domains which are also called new domains. A straight-forward solution is fine-tuning the models following the schematic diagram given in Fig. 1(a). But fine-tuning on images from additional domains leads to catastrophic forgetting, namely the models will perform badly on original domains. Joint training is effective to battle against this problem, but it has to be ensured that images from all the domains are accessible at the same time as shown in Fig. 1(b), which is unrealistic in most scenarios. Continual learning aims to gradually learn a model as the image domain changes as in Fig. 1(c). The effectiveness of the model in original domains is well maintained without accessing original images. Nevertheless, the continual learning paradigm still needs to meet the challenge of catastrophic forgetting of learned knowledge on original domains.
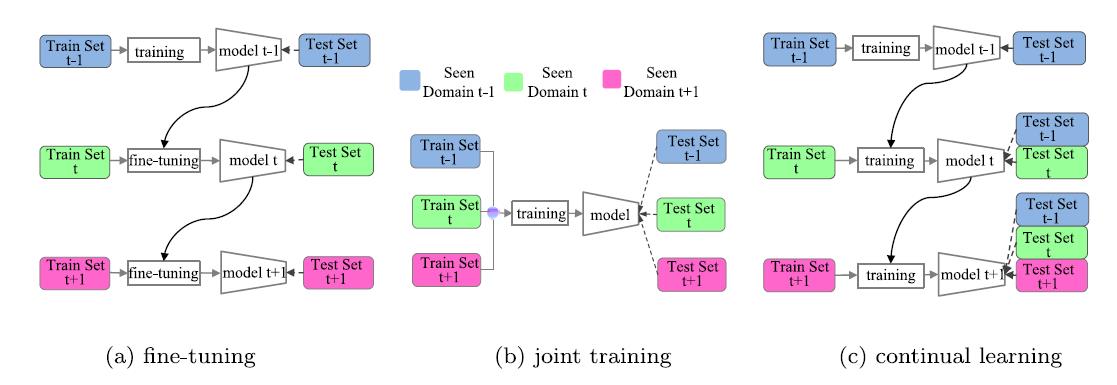
Our Contributions
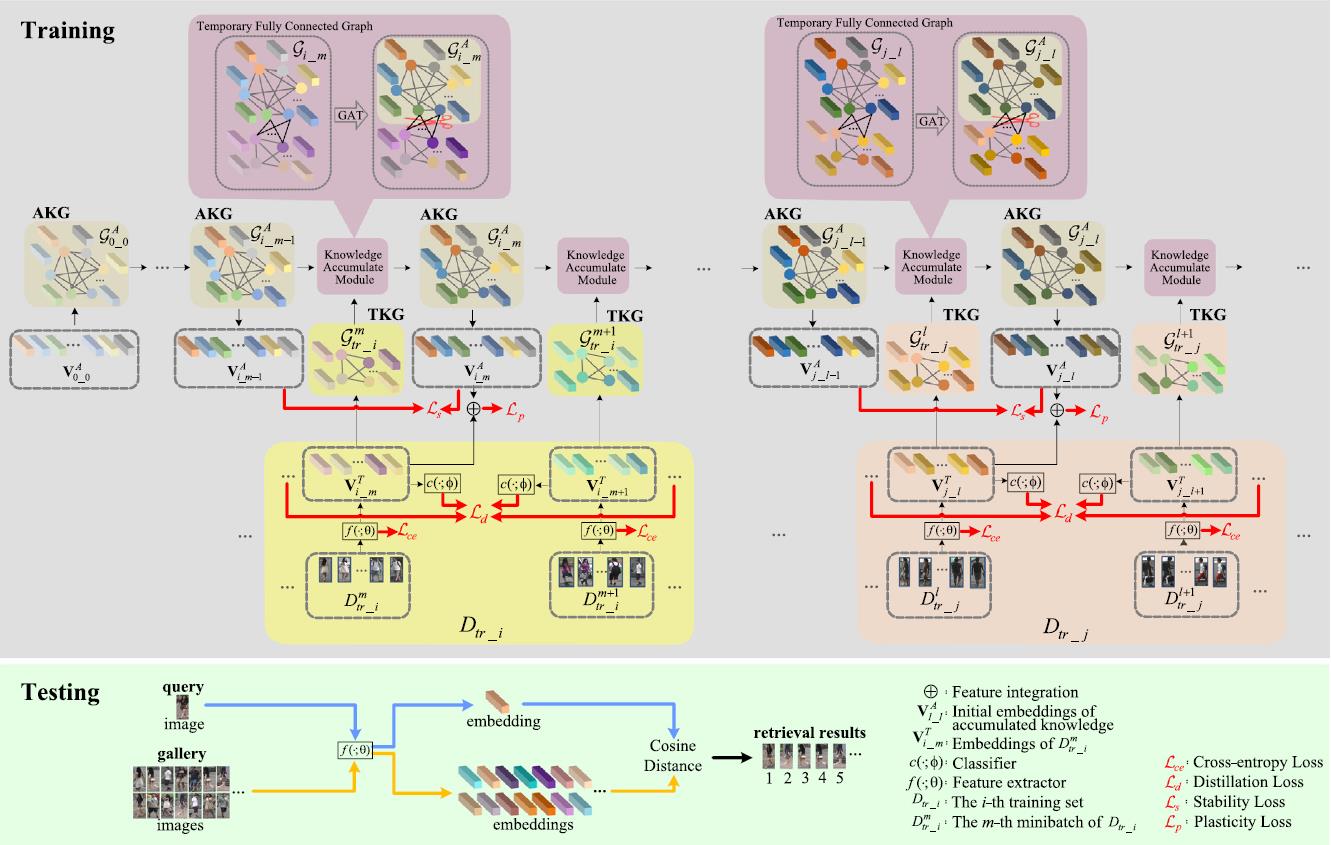
To address the above challenges, we propose a Continual person re-identification model via a Knowledge-Preserving (CKP) mechanism. Our contributions are summarized as follows:
A fully connected graph is constructed to preserve knowledge extracted from continual learning process and used to guide training.
A temporary graph, which is also fully connected, is constructed by features extracted from any given training batch.
The most related knowledge is propagated from the temporary graph to the knowledge preserving graph via a Graph Attention Network (GAT).
Experiments
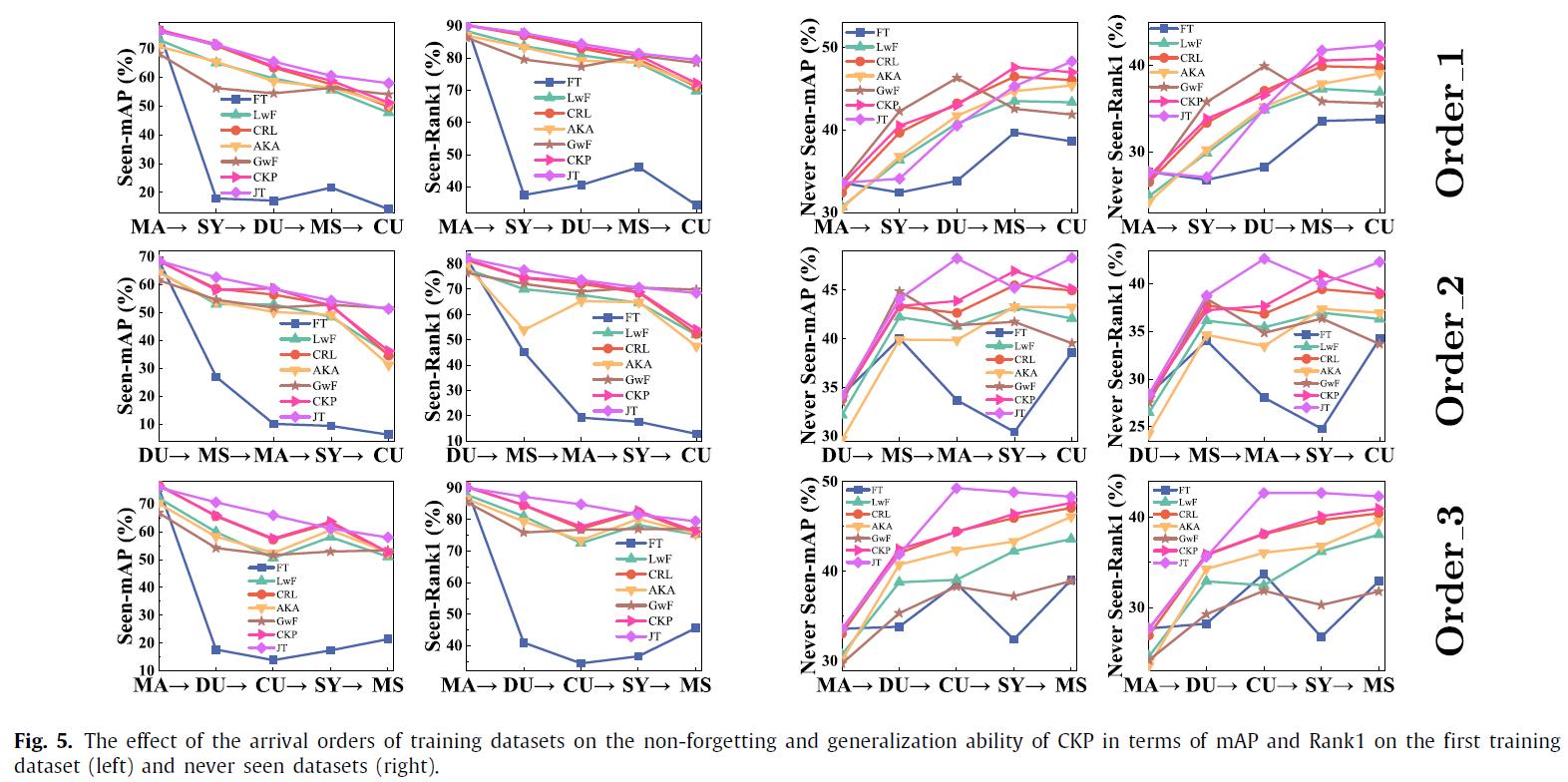
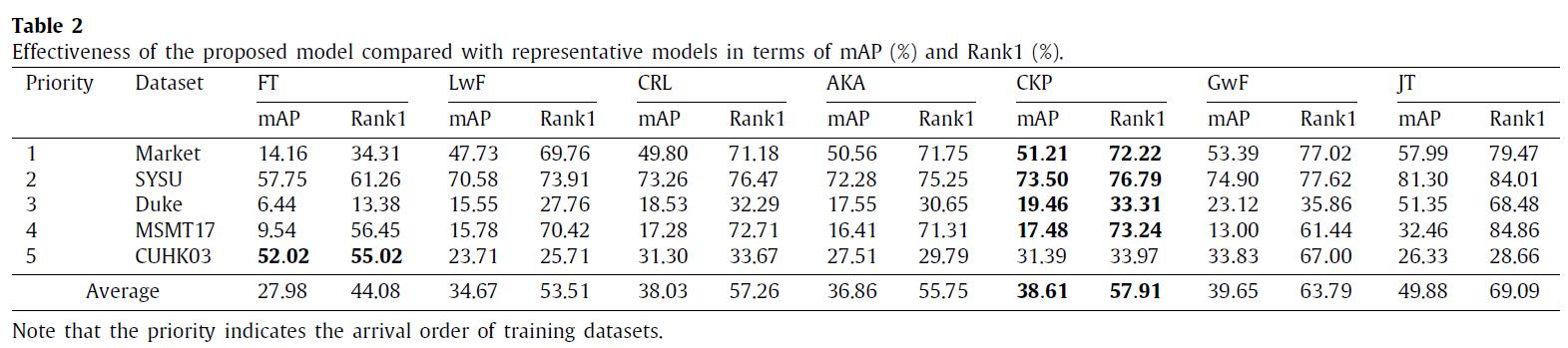
We have conducted experiments on 12 benchmark datasets of person re-identification (Market1501, DukeMTMC, CUHK03, CUHK-SYSU, MSMT17, GRID, SenseReID, CUHK01, CUHK02, VIPER, iLIDS, and PRID). Datasets are downloaded from Torchreid_Dataset_Doc and DualNorm. The comparative models include Fine-Tune (FT), Learning without Forgetting (LwF), Continual Representation Learning (CRL), Adaptive Knowledge Accumulation (AKA), Generalizing without Forgetting (GwF) and Joint Training (JT). We adopt commonly used evaluation metrics, namely the Rank-1 index and the mean average precision (mAP) to evaluate the performance of our CKP.
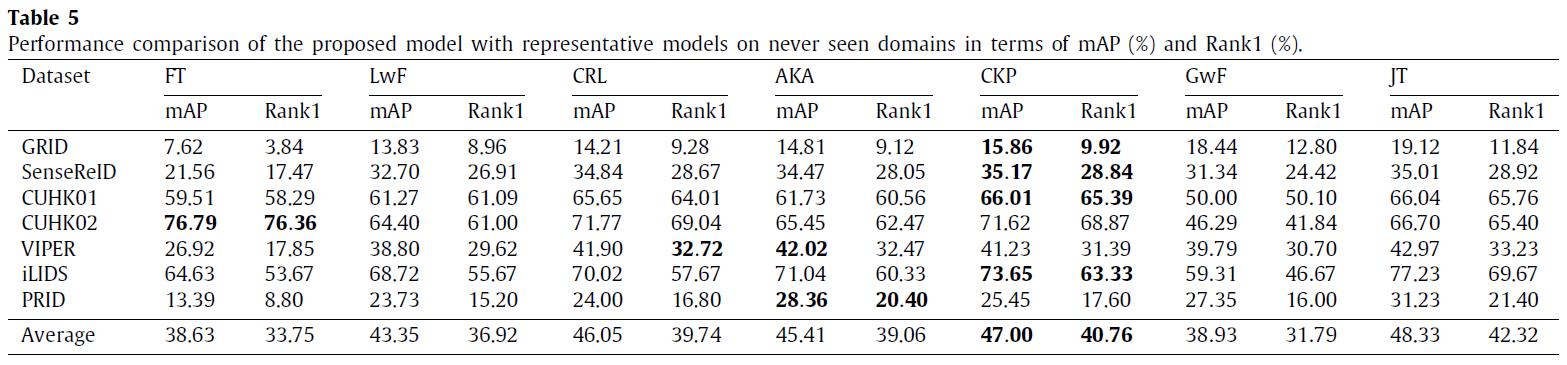
Sources
More detials please see our paper.
The code is available at ContinualReID.
Citation: The author who uses this code is defaultly considered as agreeing to cite the following reference @article{liu2023knowledge, title={Knowledge-Preserving continual person re-identification using Graph Attention Network}, author={Liu, Zhaoshuo and Feng, Chaolu and Chen, Shuaizheng and Hu, Jun}, journal={Neural Networks}, volume={161}, pages={105--115}, year={2023}, publisher={Elsevier} }

GCReID: Generalized continual person re-identification via meta learning and knowledge accumulation
Background
Person re-identification (ReID) has made good progress in stationary domains. The ReID model must be retrained to adapt to new scenarios (domains) as they emerge unexpectedly, which leads to catastrophic forgetting. Continual learning trains the model in the order of domain emergence to alleviate catastrophic forgetting.
However, continual person ReID performs terribly if it is directly applied to unseen scenarios as shown in Fig. 1. That is to say, its generalization ability is limited due to distributional differences between domains participating in training and domains not. Domain adaptation and domain generalization are helpful for continual person ReID improving its performance on unseen domains. As shown in Fig. 2(a), the standard domain adaptation requires a subset of unseen domains to participate in training to guarantee a better adaptation on the unseen domains. Domain generalization requires multiple domains to participate in training to enhance generalization. That is to say, as shown in Fig. 2(b), source domains have to be accessible in advance at the same time.
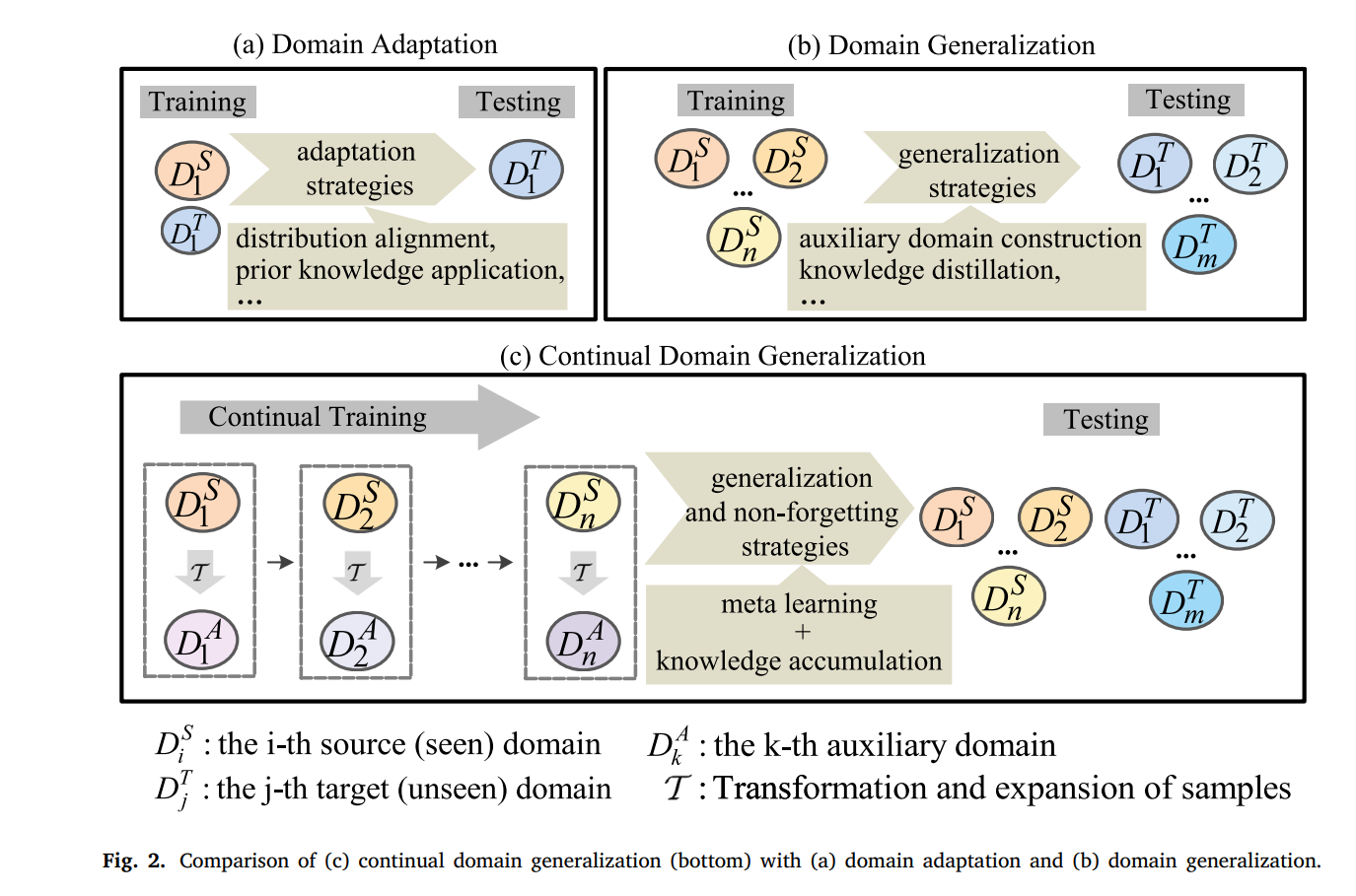
In most cases, scenarios change in order, i.e, source domains arrive with priority as shown in Fig. 2(c) rather than being accessible simultaneously. Continual domain generalization in Fig. 2(c) has to consider not only catastrophic forgetting under the continual learning paradigm (domains arrive with priority), but also the generalizability on unseen domains. Therefore, similar to resisting forgetting existing in a representative of continual learning, meta learning based parameter regularization has been introduced into this field to improve generalization of the model.
Our ContributionsIn this paper, we enhance generalization of continual person ReID for the aspect of sample diversity and distribution difference. Our main contributions are summarized as follows.
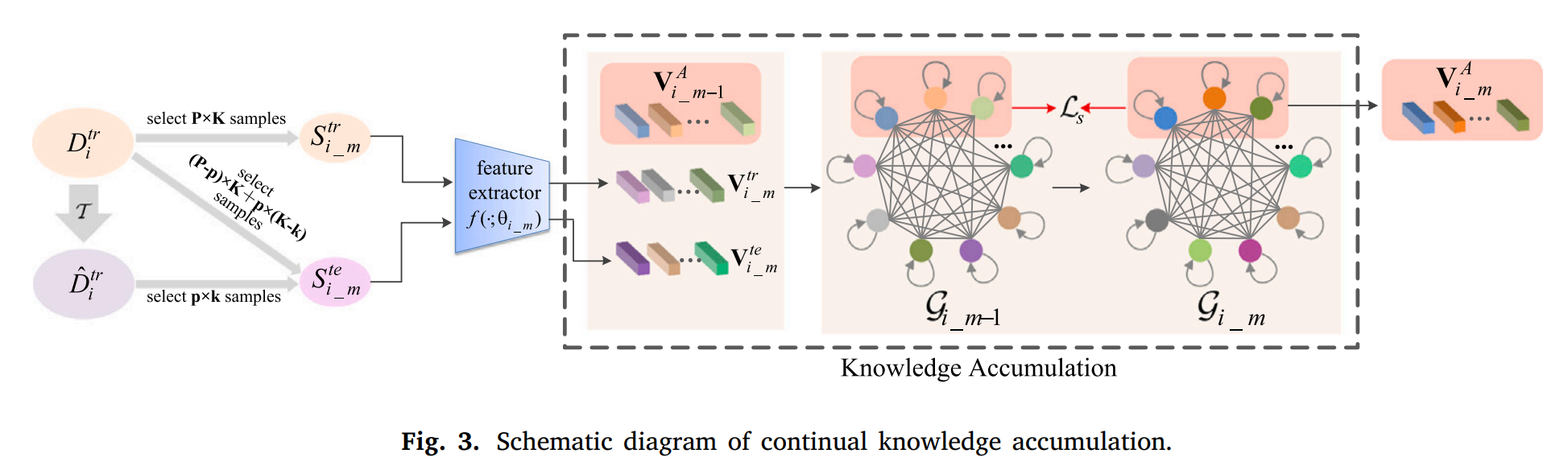
We simulate unseen domains according to priori to enhance sample diversity.
A fully connected graph is proposed to store accumulated knowledge learned from all seen domains and the simulated domains.
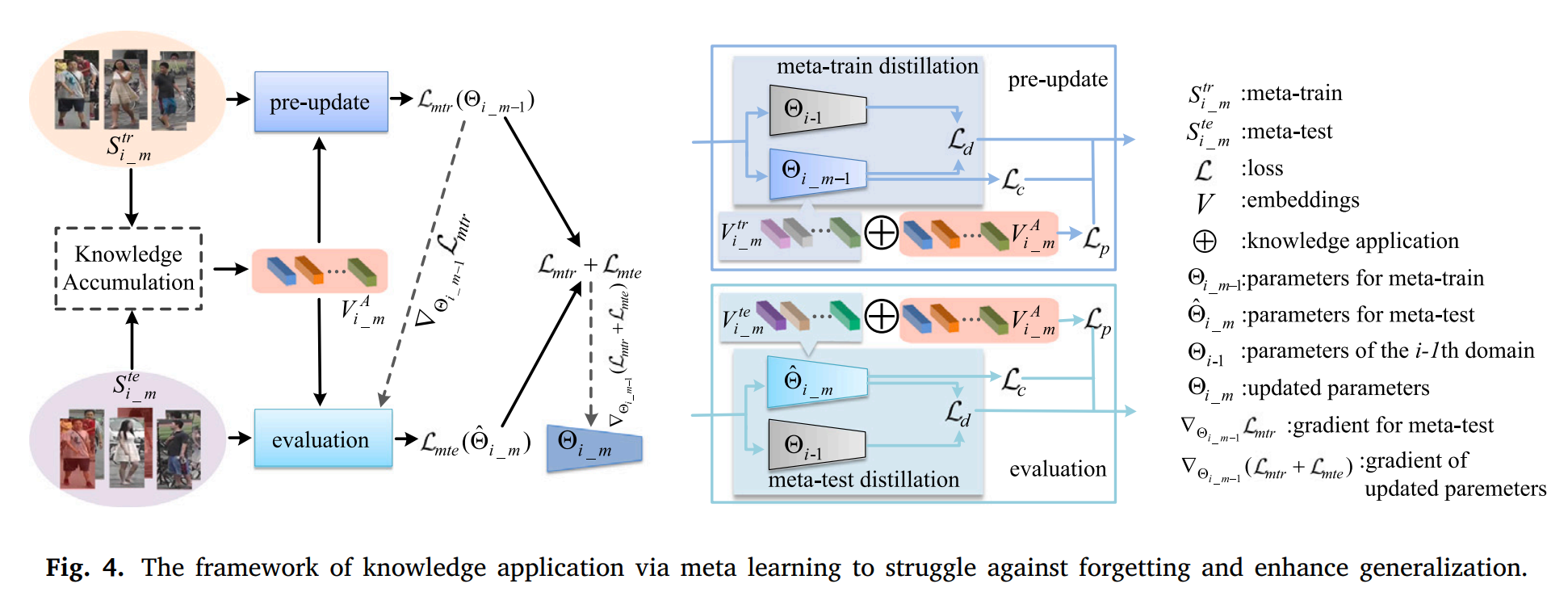
Meta-train is used to extract new knowledge from the current domain.
Meta-test is used to extract potential knowledge from unseen domains which is simulated according to priori.
The above knowledge are gathered together to update accumulated knowledge via the graph attention network.
We evaluate the proposed method and compare it with 6 representative methods on 12 benchmark datasets.
We conduct experiments on 12 person ReID benchmarks, namely Market, CUHKSYSU, DukeMTMC-reID, MSMT17, CUHK03, Grid, SenseReID, CUHK01, CUHK02, VIPER, iLIDS, the mean average precision (mAP) and Rank1 are used to evaluate performance on the datasets. Datasets are downloaded from Torchreid_Dataset_Doc and DualNorm. We compare the proposed model GCReID with 7 methods, namely Fine-Tuning (FT), Learning without forgetting (LwF), continual representation learning(CRL), adaptive knowledge accumulation(AKA), continual knowledge preserving (CKP), generalizing without forgetting (GwF), and memory-based multi-source meta-learning (M3L).
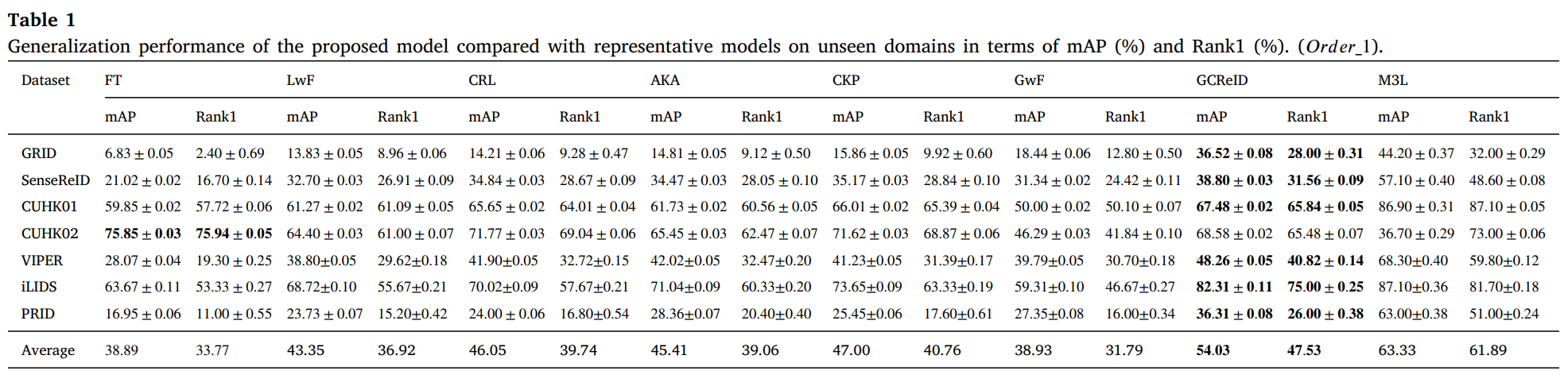
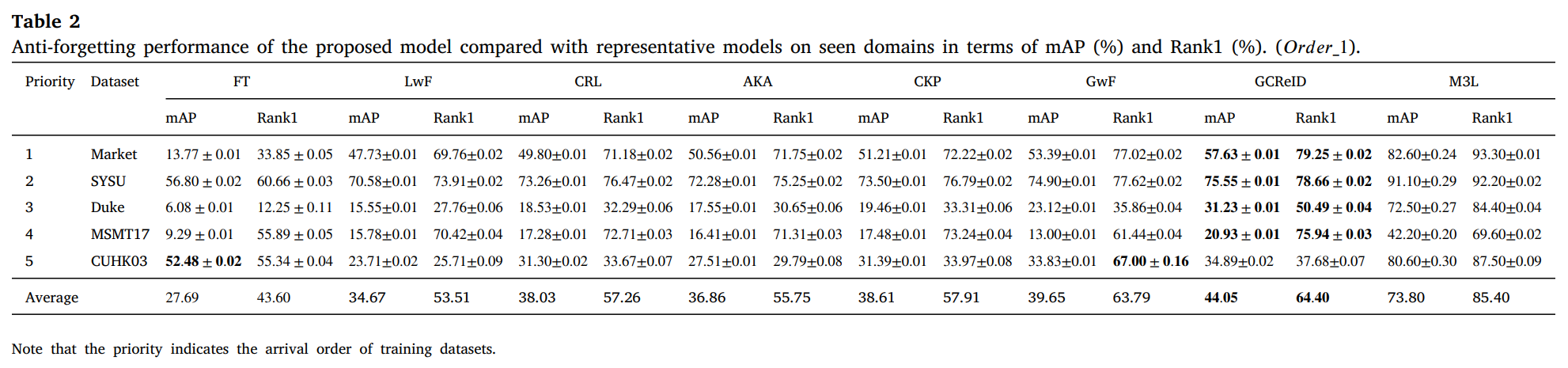
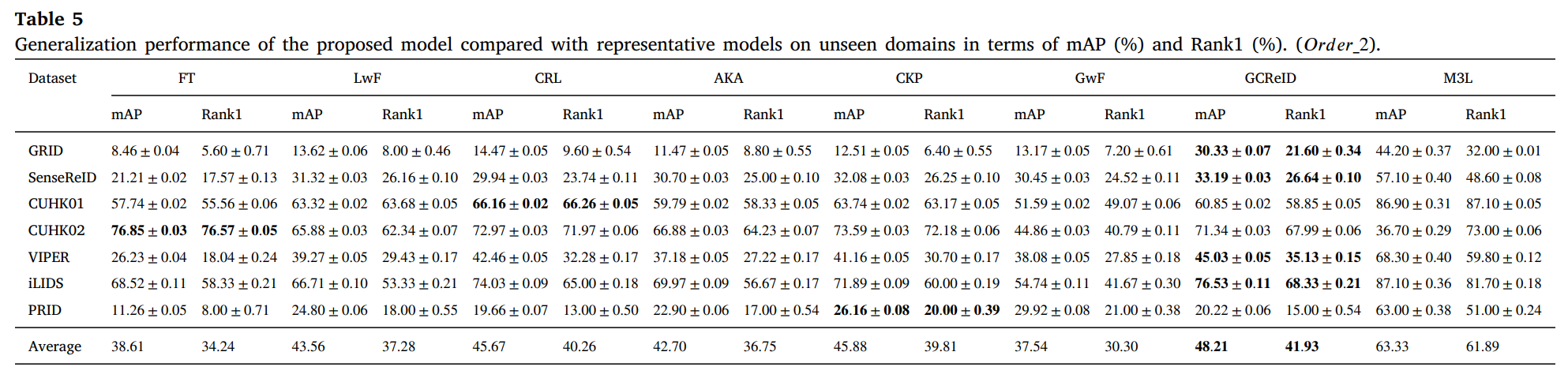
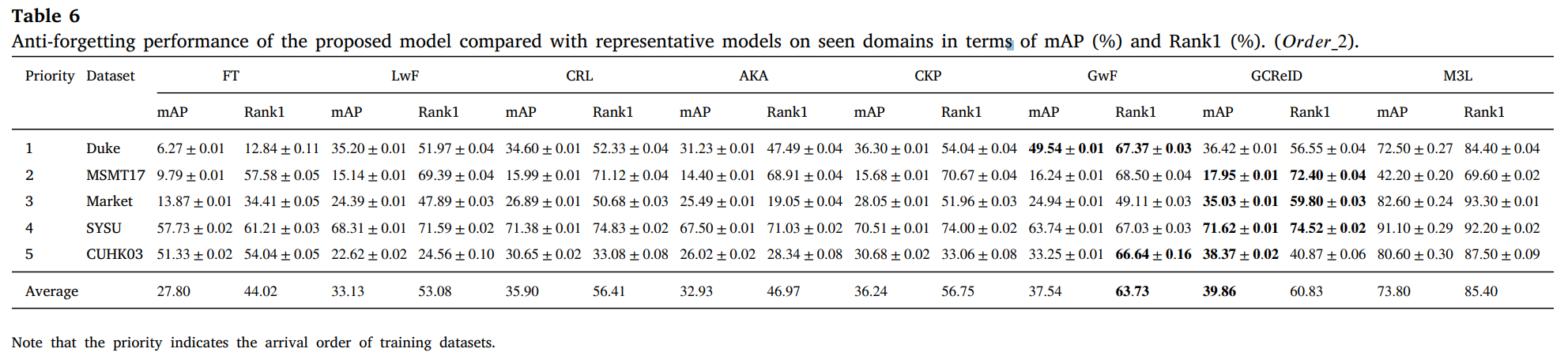
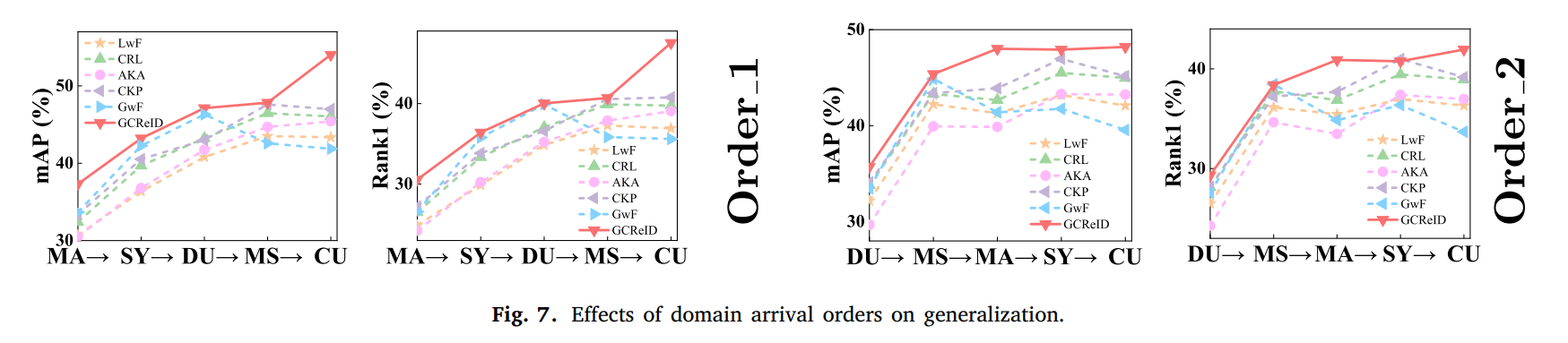
More detials please see our paper.
The code is available at GCReID.
Citation: The author who uses this code is defaultly considered as agreeing to cite the following reference @article{liu2024gcreid, title={GCReID: Generalized continual person re-identification via meta learning and knowledge accumulation}, author={Liu, Zhaoshuo and Feng, Chaolu and Yu, Kun and Hu, Jun and Yang, Jinzhu}, journal={Neural Networks}, volume={179}, pages={106561}, year={2024}, publisher={Elsevier} } }
扫描查看移动版
校址:辽宁省沈阳市和平区文化路三巷11号 | 邮编:110819